Learn about research in the Moore Lab
The work in our lab uses the tools of synthetic medicinal chemistry and chemical biology to develop new tools for studying therapeutically important molecular interactions, with a focus on protein-protein interactions. Our work currently focuses on several different therapeutic targets. Click below to find out more.
Current Projects
ER/CoA Stapled Peptides

The nuclear receptor family of transcription factors comprises 48 members and, together, they make up one of the most important classes of drug targets, particularly with respect to targeted cancer therapy. Binding of coregulators to nuclear receptors is a molecular event that modulates the expression of genes under the control of the transcription factor.
We are developing cell-permeable, peptidic molecular probes that inhibit the interactions between nuclear receptors and coactivators, and we study these interactions using fluorescence-based assays. The interaction between many of the nuclear receptors and coactivators occurs over two turns of an α-helix, using an ILXXLL motif (I = isoleucine, L = leucine, and X = any amino acid) from the coactivator.
Project team members: Changfeng Cheng
Collaborators: Jonna Frasor (UIC), Sean Fanning (Loyola University Chicago)
Grants:
American Cancer Society RSG-21-037-01-CDD; Moore (PI); 07/01/21 – 06/30/25; Selective antagonists of mutant estrogen receptors
Publications:
- Labeling of a Mutant Estrogen Receptor with an Affimer in a Breast Cancer Cell Line
- A cell-permeable stapled peptide inhibitor of the estrogen receptor/coactivator interaction
- Steroid Receptor/Coactivator Binding Inhibitors: An Update
- Recent Structural Advances in Constrained Helical Peptides
- Using Tumor Explants for Imaging Mass Spectrometry Visualization of Unlabeled Peptides and Small Molecules
- A ‘Cross-Stitched’ Peptide with Improved Helicity and Proteolytic Stability
- Stapled Peptides with γ-Methylated Hydrocarbon Chains for the Estrogen Receptor/Coactivator Interaction
Small molecules
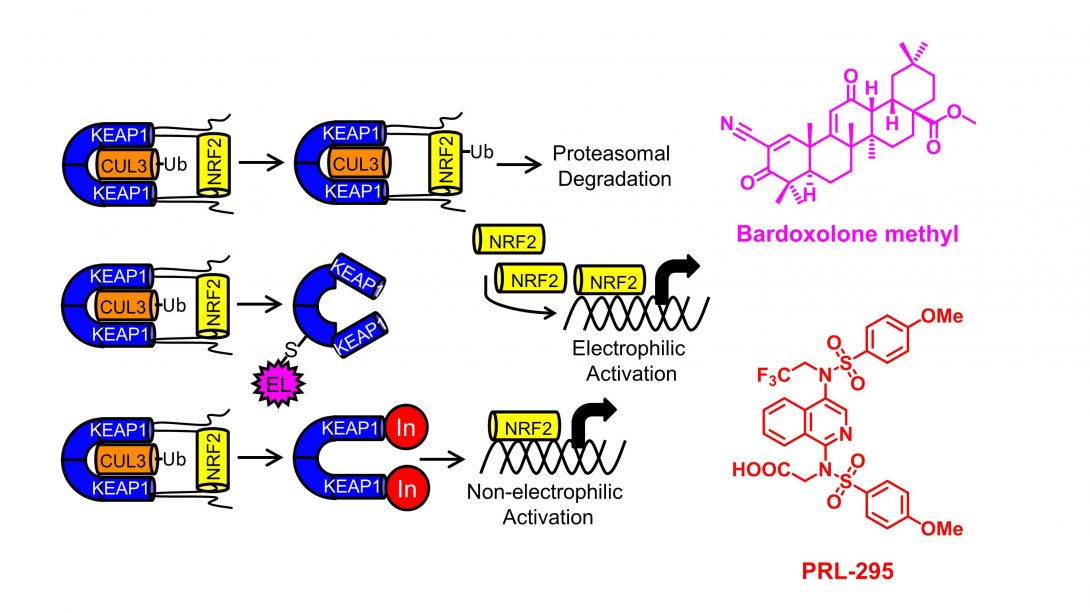
The major transcription factor in the adaptive stress response is called Nrf2; its negative regulator is Keap1, a substrate adaptor protein for a ubiquitin-ligase complex that aids in constantly degrading Nrf2. In times of electrophilic or oxidative stress, reactive cysteine residues are modified so that Nrf2 is no longer degraded. Excess Nrf2 then translocates into the nucleus and effects transcription of cytoprotective genes, especially those involved in Phase II metabolism.
There are a number of activators of Nrf2 that are known, but they are electrophilic in nature: they react with Keap1’s reactive sensor cysteines to keep Nrf2 from being degraded. We are preparing molecules that directly inhibit the interaction between Nrf2 and Keap1 as probes that can interrogate this mechanism of Nrf2 activation.
Project team members: Tatum Johnson, Shaimaa Aboukhatwa
Collaborators: Sekhar Reddy (UIC), Luisa DiPietro (UIC), Masayuki Yamamoto (Tohoku), Albena Dinkova-Kostova (Dundee)
Publications:
- Molecular basis for the disruption of Keap1-Nrf2 interaction via Hinge & Latch mechanism.
- The isoquinoline PRL-295 increases the thermostability of Keap1 and disrupts its interaction with Nrf2
- Detection of thermal shift in cellular Keap1 by protein-protein interaction inhibitors using immunoblot- and fluorescence microplate-based assays
- Synthesis and Evaluation of Non-covalent Naphthalene-Based KEAP1-NRF2 Inhibitors
- Isoquinoline Kelch-like ECH-Associated Protein 1-Nuclear Factor (Erythroid-derived 2)-like 2 (KEAP1-NRF2) Inhibitors with High Metabolic Stability
- Replacement of a Naphthalene Scaffold in Kelch-like ECH-Associated Protein 1 (KEAP1)/Nuclear factor (erythroid-derived 2)-like 2 (NRF2)Inhibitors
- Probing the Structural Requirements of Non-electrophilic Naphthalene-Based Nrf2 Activators
- Non-electrophilic modulators of the canonical Keap1/Nrf2 pathway
Apidaecin Analogs
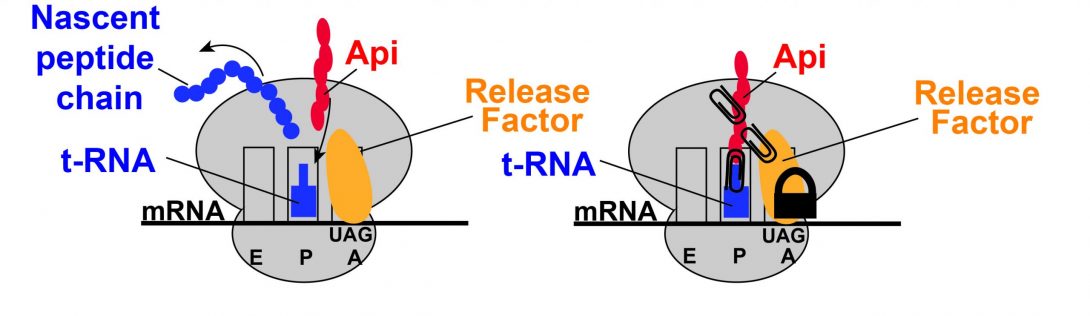
The emergence of antibiotic-resistant microbes has led to an increasing occurrence of microbial infections which are untreatable with antimicrobials currently available on the market. To keep up with the evolving microbes, it is critical to develop novel families of antibiotics with novel mechanisms of action that can be used to treat these resistant microbial infections.
Apidaecins are proline-rich antimicrobial peptides that have activity against Gram-negative bacteria. They are composed of a C-terminal region which is proposed to be responsible for antimicrobial activity and an N-terminal region which is proposed to be responsible for uptake of the molecule to the interior of the cell. The apidaecins target translation through a novel mechanism of action: the stabilization of the translation termination complex of the 70S ribosome, P-site t-RNA, and A-site release factor. In a collaboration with the lab of Shura Mankin and Nora Vázquez-Laslop at UIC, we are developing Apidaecin derivatives with optimized antimicrobial activity and proteolytic stability.
Project team members: A’Lester Allen
Collaborators: Shura Mankin (UIC), Nora Vázquez-Laslop (UIC), Yuri Polikanov (UIC)
Grants:
NIH R01 AI162961; Mankin, Moore, Polikanov, Vázquez-Laslop (MPI); 02/01/22 – 06/30/27; Advancing ribosome-targeting antibacterial peptides with a unique mechanism of action
Publications:
Sin 3 Inhibitors
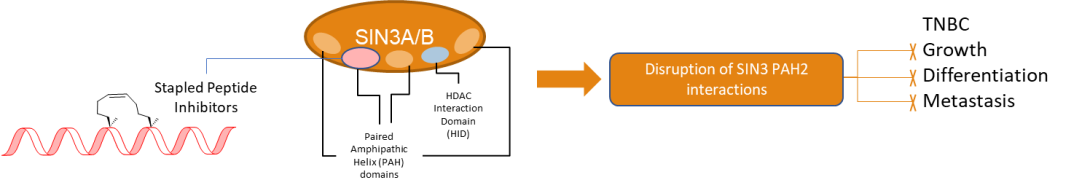
Triple Negative Breast Cancer accounts for 20% of all breast cancer cases and is one of the most clinically challenging subtypes of breast cancer to treat. Recently, Histone Deacetylase-associated transcriptional regulatory proteins SIN3A and SIN3B have emerged as potential molecular targets for Triple Negative Breast Cancer therapeutics. Peptides and small molecules targeting the PAH2 domain, a well-characterized protein-interaction domain of SIN3 homologs, have shown induction of epigenetic reprogramming along with upregulation of differentiation markers and downregulation of epithelial-to-mesenchymal transition markers in Triple Negative Breast Cancer cell lines. We are developing hydrocarbon-stapled peptides targeting the PAH2 domain with improved affinity, cell-permeability, and protease resistance.
Project Team Members: Changfeng Cheng
Collaborators: Ishwar Radhakrishnan (Northwestern), Dai Horiuchi (Northwestern)
Ebola
Viral outbreaks have and continue to persist globally, increasing the need for drug discovery and development of novel antivirals. Some of the most aggressive and life-threatening viruses belong to the Filoviridae family. One of these is Ebola virus, which expresses an enveloped glycoprotein (GP) that mediates several steps in the viral entry process. We have designed and synthesized small molecule inhibitors that bind GP and block viral entry within host cells. Our work in collaboration with the Lijun Rong lab at UIC aims to optimize the antiviral activity and drug-likeliness of our small molecule GP inhibitors as an effort to develop effective and stable Ebola antiviral treatments.
Project team members: Irina Gaisina, Destiny Durante
Collaborators: Lijun Rong (UIC)
Grants:
NIH U19 AI171954; Bannister (PI); Role: co-investigator; Midwest AViDD Center Core C: Medicinal Chemistry and DMPK; 05/16/22 – 04/30/27
NIH R41 AI126971; Rong (PI), Role: co-investigator; 08/16/19 – 07/31/22; 4-(Aminomethyl) benzamides as novel anti-Ebola agents
Influenza
Influenza is an infectious disease known to have a high morbidity and mortality rate. Many influenza A isolates have shown resistance to approved therapeutics currently being used, creating a need for therapeutics with novel mechanisms of action. Hemagglutinin (HA) is a glycoprotein located on the surface of the virus, which facilitates viral entry into the host cell. Using high-throughput screening, our group has identified small molecule entry inhibitors targeting hemagglutinin. We are currently developing multiple compound scaffolds targeting hemagglutinin in Group 1 and Group 2 influenza viruses.
Project Team Members: Irina Gaisina, John Sloan
Collaborators: Lijun Rong (UIC)
Grants:
NIH R42 AI55039; Rong (PI), Role: co-investigator; 05/05/22 – 04/30/25; Development of 4-(aroylamino) piperidine-based entry inhibitors as anti-influenza therapeutics
Photoprobes
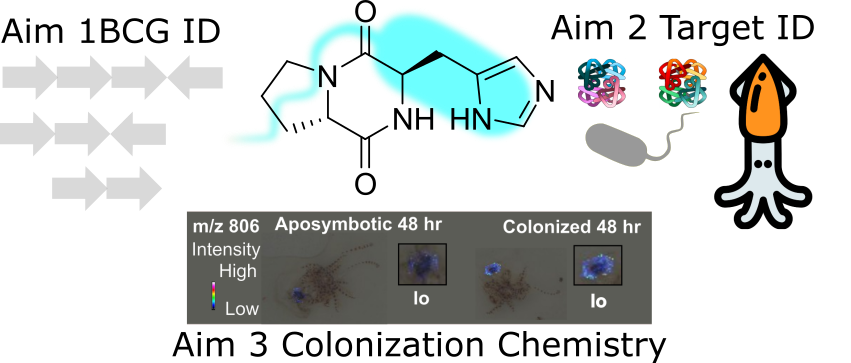
There is significant interest in identifying and characterizing small molecules that underlie host-microbe communication in plant, animal, and human host-microbiome interactions. The naturally simple association between Euprymna scolopes bobtail squid and Vibrio fischeri luminescent bacteria provides a symbiotic relationship that can be interrogated in situ to discover functional compounds. Bacterial colonization of the host is accessible to sophisticated bacterial genetic and genomic approaches, and the site of infection can be imaged in live animals. Our collaborative efforts have developed novel methods to identify and study small molecules in this symbiosis. This work has resulted in the identification of a small organic molecule, termed diketopiperazine (DKP), as a potential contributor to symbiotic biofilm and luminescence phenotypes from V. fischeri. We have used imaging mass spectrometry to identify additional biofilm-associated molecules, and, using structure elucidation and synthetic chemistry, we have synthesized large amounts of the DKP. Here, we will leverage our interdisciplinary expertise in imaging mass spectrometry, structure isolation and elucidation, forward genetic screening, and synthetic bioorganic chemistry to identify the function of this DKP and identify the biosynthetic origin of this molecule to explore whether it is used in other microbial-host systems. This model system will allow us to address current knowledge gaps in our understanding of the small molecules that symbiotic bacteria produce during colonization of host specific tissues.
Project Team Members: Shaimaa Aboukhatwa
Collaborators: Laura Sanchez (UCSC), Mark Mandel (Wisconsin)
Grants:
NSF IOS-2220512; Sanchez/Moore/Mandel (MPI); 08/15/2022 – 07/31/2025; Collaborative Research: Chemical signaling in a host-microbe symbiosis
Publications: